13 FEB 2024
Collaborative session notes https://docs.google.com/document/d/1ykqPGchur5QwnqAnE9jEGIgzZZJcCyEhb557j3j7l10/edit?usp=drive_link
Group(s) submitting the application:
Meeting objectives:
In this session, we will focus on the “FAIRification of Genomic Annotations WG”. This WG aims to establish a public infrastructure to improve the FAIRness of a particular and widely used type of life science data, namely “genomic annotations” (or “genomic tracks”). Genomic annotations are data that describe reference genome sequences from species spanning the entire tree of life. The sequences (typically DNA) are annotated with position-anchored information based on a large variety of data sources resulting from e.g. experiments, predictions, databases, and manual processes. Use cases include analytical applications in biomedical research, genome annotations for biodiversity assemblies, and data exchange infrastructures in the context of genome browsers, which is a particular type of visualisation service.
We want to use the session in the RDA Plenary to present the main use cases and goals of the working group, including examples of existing work that the WG will build on. On this basis, we will open up for a discussion on the challenges within the use cases, individual tasks from the WG work plan, and possible intersections with other RDA groups and efforts elsewhere, such as the potential adoption of existing recommendations. We want to make use of the session to start convening a community in the context of the WG. We, therefore, aim to invite broadly to the session also outside of the current RDA community, e.g. from other entities such as ELIXIR and the Global Alliance for Global Health (GA4GH).
Meeting agenda:
- Welcome and a brief introduction to the working group (5 min)
- Lightning talks on use cases and previous work (30 mins)
- Assoc. Prof. Anna Bernasconi (Politecnico di Milano)
- Dr. Sveinung Gundersen (ELIXIR Norway, University of Oslo)
- Assoc. Prof. Nathan Sheffield (University of Virginia)
- Dr. Adam Wright (Ontario Institute for Cancer Research)
- Dr. Thomas Brown (Leibniz Institute for Zoo and Wildlife Research)
- Overview of the WG work plan (10 mins)
- Open discussion with a focus on challenges within the use cases, individual tasks from the WG work plan, and possible intersections with other RDA groups and efforts elsewhere (35 mins)
- Wrap-up (5 mins)
Target Audience:
Anyone interested – the talks will not assume domain knowledge. The WG aims to build a broad community of individuals and groups with a passion for data integration related to genomic annotations, encompassing data producers, domain experts, tool/service developers, FAIR/RDM specialists, analytical end users, and others not falling into any of these categories.
Group chair serving as contact person:
Brief introduction describing the activities and scope of the group:
The FAIRification of Genomic Annotations Working Group (WG) will focus on the challenges of harmonising metadata and software solutions to improve the discovery and reuse of publicly available “genomic annotation” data. The WG is one of the pilots to receive support from the RDA TIGER project.
Genomic annotations
Since the completion of the Human Genome Project, we have witnessed an explosion of publicly available datasets describing e.g., aspects of functional processes that relate to particular locations along reference DNA sequences. Genomic annotations, sometimes referred to as “genomic tracks”, refer to data files that annotate genome reference sequence positions and can be visualised in genome browsers (see Figure 1), such as the UCSC Genome Browser and Ensembl, or analysed non-visually using computational tools that span the domains of life science (e.g., statistical colocalization analyses).
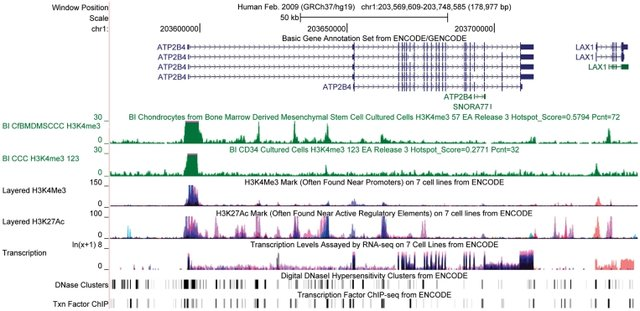
Figure 1: Example of tracks from the ENCODE consortium imported into the UCSC Genome Browser. From Rosenbloom et. al. (2011). ENCODE whole-genome data in the UCSC genome browser: update 2012. Nucleic acids research. 40. D912-7. 10.1093/nar/gkr1012. License: CC BY-NC 3.0
Goals of the working group
The WG will establish a minimal metadata schema based on the harmonisation of existing schemas and recommendations, and gradually improve this schema as required by a set of use cases. To foster the availability of metadata that adheres to the schema, the WG will develop practical recommendations and adopt and improve existing data flow infrastructures to build scalable and maintainable metadata transformation flows from multiple data sources. Recommendations will be developed for publishing and registering the harmonised metadata – with persistent and globally unique identifiers – so that it can be harvested by services that improve the discovery and repurposing of the metadata (and associated datasets). Lastly, the WG will develop a harmonised API for making use of search and discovery services by downstream users and tools.
The main use cases to be considered are:
- Discovery of genomic annotations through harmonised metadata to improve the availability and scope of datasets for analytical applications, including methodology-oriented software tools and AI/machine learning efforts. A main focus area will be human biomedical research, but this use case will also include support for research on other species in alignment with the interests of the WG contributors.
- Establish genome annotations for biodiversity assemblies as FAIR objects with improved metadata, integrating with relevant infrastructure and tools, such as for automatic or manual annotation of genome assemblies.
- Enhance genome browser-related infrastructure for track hubs with harmonised metadata and related services, to simplify the process of generating, hosting and registering metadata-enriched track hubs, improve discovery of track hub-hosted data at the level of individual tracks, and facilitate the integration of metadata-enriched track hubs with genome browsers and other analysis tools.
The WG aims to build a broad community of individuals and groups with a passion for data integration related to genomic annotations, encompassing data producers, domain experts, tool/service developers, FAIR/RDM specialists, analytical end users, and others. The long-term goal is to build a sustainable infrastructure that improves the FAIRness of genomic annotations, with a particular focus on improving the end-user experience.
Short Group Status:
Since our previous BoF session in the International Data Week / 21st RDA Plenary, we have focused on raising interest and finalising the case statement. There are now four confirmed co-chairs of the initiative. We believe we have established sufficient interest to move forward and apply to become a working group. The case statement will be submitted within a few weeks of submission of this workshop application. We assume the process of establishing the working group has been completed by the time of the plenary and that we have also started regular meetings by then.
Type of Meeting:
Working meeting
Additional links to informative material:
- Case statement of the “FAIRification of Genomic Tracks WG”
- Slides and video from the BoF session in the International Data Week / 21st RDA Plenary
- Blog post about the FAIRtracks project, which took the initiative of starting the WG
- The FAIRtracks website contains background information on genomic tracks (annotations), relevant FAIR-enabling solutions, the FAIRtracks metadata schema, and more.
Avoid conflict with the following group (1):
Avoid conflict with the following group (2):
Meeting presenters:
Anna Bernasconi*, Sveinung Gundersen*, Ryan O’Connor, Nathan Sheffield*, Adam Wright*, and a representative from the biodiversity community (TBD). * WG co-chairs
Are you willing to host a second, repeat, session at a different time zone?:
Yes
