Innovating Tools To Create A Future For Biodiversity Genomic Data Governance To Support A New Era Of Ethical Genomic Research Practitioners With The Help Of EOSC Future And RDA
By Ann Mc Cartney, 23 February 2023

The effort of creating a catalog of Earth’s biodiversity, known as the European Reference Genome Atlas (ERGA) is accompanied by the project “Contextual Metadata Futures: Building Indigenous Data Provenance Capacity for the European Reference Genome Atlas” funded by the EOSC Future and RDA. As a result of this work, we strongly suggest that inside of EOSC the CARE principles should be made more visible and a general awareness should be created. We contributed to this during the project and it needs to be continued in EOSC advisory groups, in the RDA ambassadors’ program and finally in everyday practice of science. As a westernized project co-ordinated by white privileged researchers, ERGA-Pilot sought out opportunities to incorporate sovereignty in a way that implemented creative and innovative tools already being produced by leading Indigenous Data Sovereignty experts. For this, a partnership with Global Indigenous Data Alliance (GIDA) and Local Contexts was established. The partnership focused on developing novel strategies to operationalize the CARE principles for Indigenous data governance over samples and data generated for the purposes of reference genome production that had been collected from IPLC lands, air, and waters. Whilst evaluating the project design, it was determined that integrating the Local Contexts Traditional Knowledge and Biocultural Label and Notices system within the ERGA-Pilot as a standard of practice, would ensure that any Indigenous rights and interests could be disclosed and associated with all IPLC samples and data collected in partnership with ERGA.
In the midst of our sixth mass extinction, according to recent reports from IUCN a total of 41,000 of our known species are threatened with extinction. The extinction of Earth’s species is not confined to a particular taxonomic group, nor to a specific geographic area (Fig 1), but rather it is a global problem that has been considered by the Convention on Biological Diversity to be a “common concern” of all humanity. Despite the conservation and preservation of our biodiversity being a shared global problem, the solutions for each species, ecosystem, and geographic location will need to be tailored accordingly. Targeted universalism is one strategy proposed to achieve a shared mission through locally contextualized solutions.
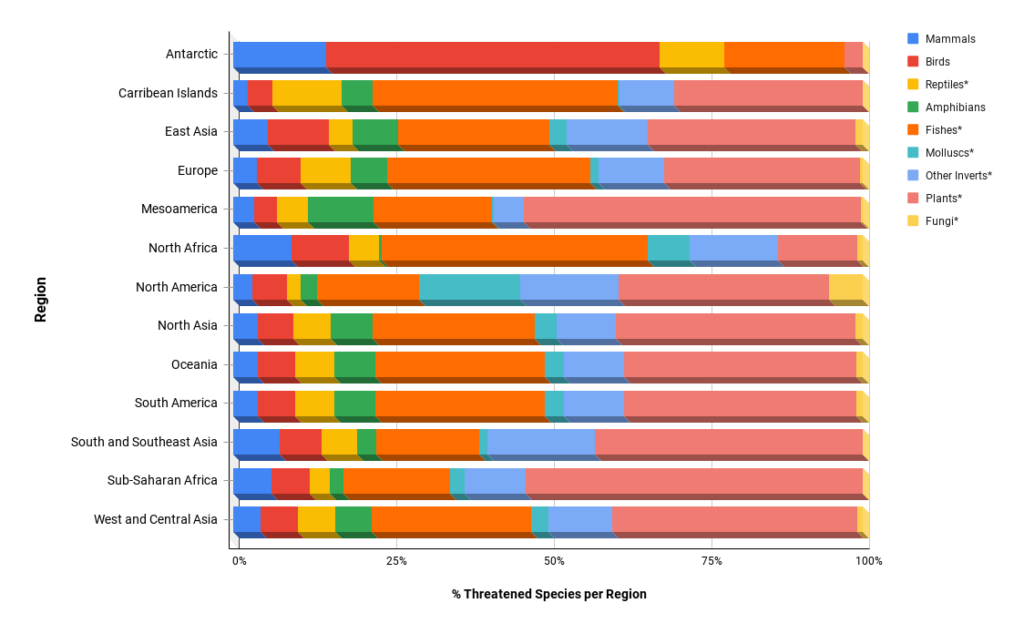
Fig 1: Threatened species by taxa per region. Data based on IUCN.
Genomes for conservation
A key aim of the biodiversity genomics research enterprise is to provide a catalog of Earth’s biodiversity through genomic sequencing technologies, using this information to develop tools, metrics and indicators that can contribute to the conservation and preservation of species. Acknowledging the magnitude of work ahead, in 2018 the Earth BioGenome Project (EBP) was established. The EBP as an umbrella organization acts to synergize the efforts of biodiversity genomic initiatives across the globe. The organization is growing, with >50 affiliated initiatives to date. In 2021, the first regional node of the EBP was established, the European Reference Genome Atlas (ERGA).
The mission of ERGA is to use DNA sequences to catalog and build our understanding of all Europe’s eukaryotes. For each species, a vast quantity of sequencing information will be produced – enough to cover the entire DNA record, or genome, of the species. This sequence information will then be carefully placed together, akin to placing the pieces of a jigsaw puzzle together. However, as there are typically many many more DNA sequences than jigsaw puzzle pieces, this is done through computational algorithms to ensure the sequences are put together in the correct order. After assembling the pieces of the sequence puzzle – the product is called a reference genome.
Reference genomes are powerful tools yielding the ability to explore previously unknown aspects of the species we share our planet with. Genomes from the same species can be compared to understand the diversity within – an indicator of a species resilience to climate changes. Genomes from different species can be compared to better understand their evolutionary relationship, providing a clearer understanding of the branches that compose the tree of life. Genomes also contain genes that can be compared to identify genes that are associated with specific characteristics and can have huge implications for biodiversity and ecosystem health (including humans!), food-security, and ecosystem services. The breadth of applications made possible by reference genomes highlight the importance of investing resources into their generation for all species.
Situating genomes in a socio-political context
The creation of genomes is not outside of the socio-political realities that exist across the globe today. Generating data to create accurate and complete reference genomes requires expensive sequencing equipment, laboratory access, a skilled workforce, and significant computational resources. Just like the distribution of biodiversity, the resources to create reference genomes are not evenly distributed across the globe. For ERGA specifically, this is evident if one looks at the OECD reports on GDP per capita and the percentage investment in R&D per country in Europe. To address this and learn about how these structured inequities would manifest in ERGA, a Pilot Project (ERGA-Pilot) was established. From its outset, ERGA-Pilot recognised that the purposeful inclusion of segments of the population, Peoples, and communities that have and continue to be left outside of research was fundamental to the long-term success of the ERGA. The Project was also cognisant that the foundations built during the pilot phase would have huge implications on who was included, had access to, and benefitted from the production of genomes across Europe into the future. To this end, ERGA-Pilot undertook a critical evaluation of justice, equity, diversity and inclusion (JEDI) throughout all stages of the decision-making processes associated with the project design. Here, the intentional acknowledgement, recognition and participation of Indigenous Peoples and Local Communities (IPLCs) and respect for their sovereignty was considered a priority.
By intertwining JEDI into the scientific mission of the ERGA-Pilot in this way, a strategy for establishing a decentralized, accessible, and scalable infrastructure that supported the production of reference genomes for all species, and was accessible to all researchers across Europe was designed. This infrastructure was designed to ensure that it was responsive to the rights codified in the United Nations Declaration on the Rights of Indigenous Peoples (UNDRIP), that outlines Indigenous Peoples’ right to exercise sovereignty over their genetic resources and data.
Contemporising genome data governance for a more just, equitable and inclusive genomic future
IPLCs have, and continue to be left outside of the research enterprise, specifically genomic research. Typically, when genomic research has been conducted with IPLCs, it is usually done on them, their knowledge systems, and resources for the benefit of an external project’s research agenda. Seldom does the research provide meaningful results back to the IPLC, and rarely does it sustainably include IPLCs as equal partners. IPLCs have grown tired of “gifting” their time, resources, and knowledge to research under the auspices of the “public-good” without involvement, benefits, and fair attribution. Many IPLCs are now taking this into their own hands. Indigenous digital technologies, data governance principles, and research organizations are being created to take agency over Indigenous research, samples and data. These processes and procedures are driven by Indigenous ways of knowing creating a new research ecosystem that responds to contemporary research needs but remains outside of, and free from colonial legacies.
Contextualising Genomes
The power of reference genomes is driven by the breadth of potential research applications it can support. In 2016, the FAIR principles were developed to provide researchers data-centric guidance for realizing the full potential of data, for their own research purposes and for secondary users. The principles have since become a dogma within the biodiversity genomics research community, and an almost equivalent value is now being placed on the contextual information associated with generating the genome data – the metadata. Similar to many research standards of practice, existing metadata standards such as Darwin Core are plagued by colonial legacies. This has resulted in the adoption of standardized metadata schemas that have prioritized the collection of information that has been deemed important to western and white researchers. Subsequently leading to the standardized erasure of information of importance to IPLCs from sample and data records.
Local Contexts is an Indigenous led organization dedicated to rectifying this wrong through the development of a human- and machine- readable disclosure system that functions to streamline the inclusion of Indigenous permissions, protocols and provenance into metadata records, within digital environments. The method of disclosure is initialized by both the researcher and IPLC registering on the Local Context Hub. From here, the researcher can create a research project for the samples or data and assign a Biocultural Notice to the project. This Notice will be immediately sent through the Hub to the partnering IPLC. Upon receiving the Notice, the IPLC can validate the Notice and issue a customized Traditional Knowledge and/or Biocultural Label/s disclosing any provenance, protocol or permission information the community would like to associate with the project and their resources. Each registered project contains a unique permanent identifier that can be placed into the metadata record of the associated samples and data, providing a long-term link to the disclosed Indigenous interests.
For ERGA-Pilot, generating genomes for European species by researchers across all Europe required the development of a comprehensive and robust metadata collection procedure that could ensure that all samples collected and data generated, were associated with all relevant contextual information. Here, an ERGA metadata schema was established along with a supporting standard operating procedure. The completion of a valid metadata schema was mandatory for participation in the ERGA-Pilot. Metadata completeness and validity for the project was inspected by COPO, a metadata brokering platform. During schema development, a new, controlled, and validatible field was generated to support the implementation of the Label and Notices’ unique permanent identifiers (Fig.2). A guidance document was also developed to offer support for researchers new to the Label and Notices system.
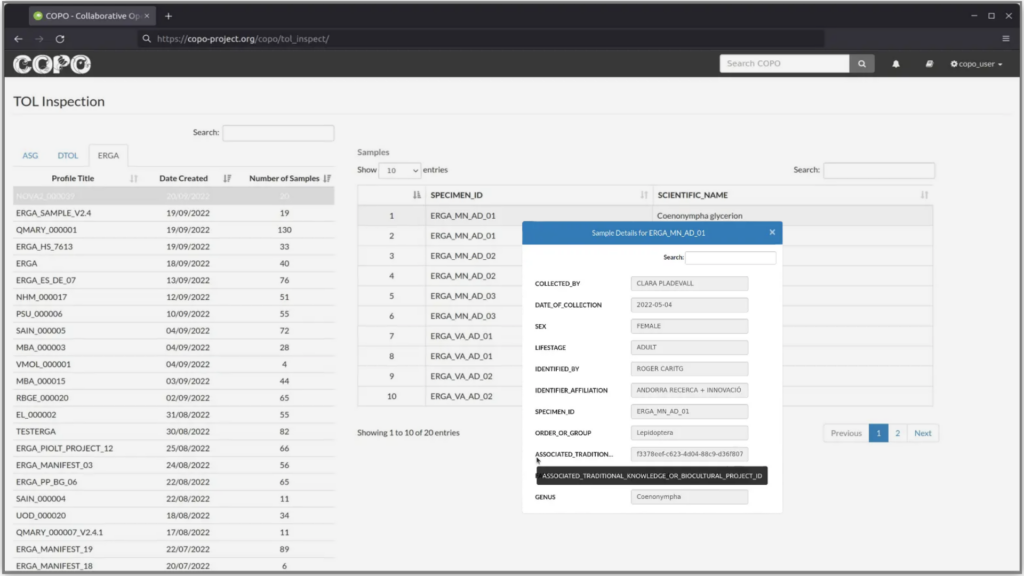
Fig. 2: Graphical representation of the unique project ID or UID implementation in COPO after user metadata manifest upload.
Decolonising metadata schema across biodiversity genomic research
Embedding Indigenous rights and interests as part of the ERGA-Pilot metadata collection standard of practice provided every participating researcher the opportunity to collect samples and generate genome data that are in alignment with both the FAIR and CARE principles. Making space for Indigenous rights and interests as a standard of practice during the metadata collection procedure in this way has consequently resulted in the inclusion of these fields into ERGA as it moves beyond its Pilot Phase. However, we hope that by utilizing the ERGA-Pilot as a regulatory sandbox showcasing the benefit of this approach, that it will motivate and inspire uptake by other biodiversity genomics initiatives, metadata standards, public digital repositories, and scientific journals to build a more just, equitable and inclusive future for the scientific research enterprise at large.
